What is SPSP?
SPSP in a nutshell
SPSP is the national genomic platform to support pathogen surveillance programmes, real-time monitoring of outbreaks and of circulating strains. It contributes to reduce the socio-economic burden of infectious diseases and protect health in a One Health perspective.
It offers a secure infrastructure that enables timely, coordinated, and science-based responses to infectious disease challenges. It facilitates controlled, real-time sharing of microbial genomic data alongside structured clinical, epidemiological, and environmental metadata—ensuring early detection and targeted intervention.
SPSP is built on the One Health approach, recognizing the deep interconnection between human, animal, and environmental health. By integrating genomic data from clinical, veterinary, foodborne, and environmental sources, the platform provides a comprehensive view of how pathogens emerge, evolve, and spread across ecosystems.
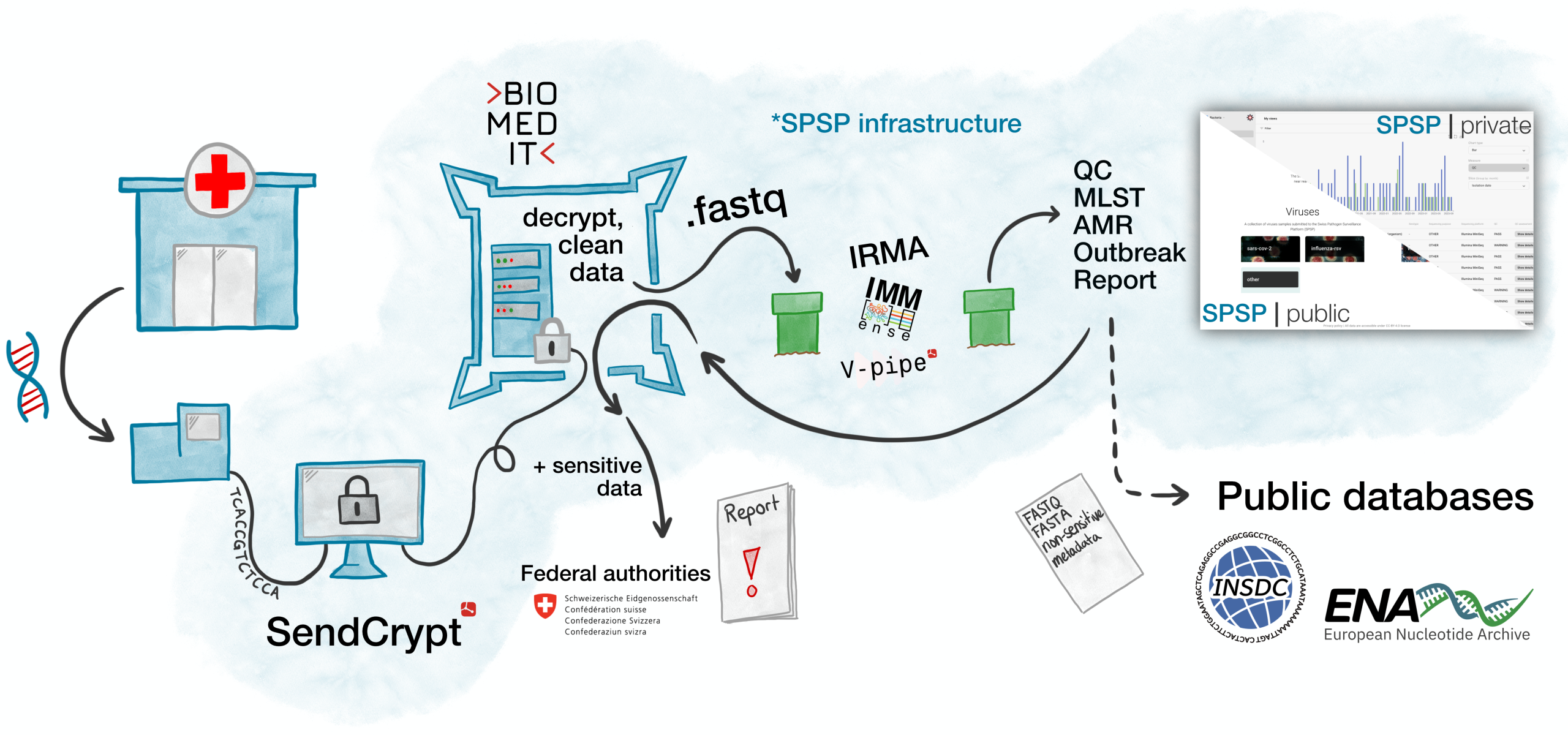
Hosted at the SIB Swiss Institute of Bioinformatics, and operated in collaboration with university hospitals and academic institutions in Basel, Lausanne, Geneva, Bern, and Zurich, together they provide a unified framework to support surveillance, policy planning, and infectious disease research.
Its governance model ensures secure and ethical data use, based on a robust legal framework including a Consortium Agreement and SPSP Ethical Approval. SPSP complies with SPHN data security standards and adheres to FAIR principles (Findable, Accessible, Interoperable, and Reusable) enabling responsible use of data for both public health surveillance and infectious disease research at national and international levels.
Benefits for public health authorities
“During the pandemic, SPSP has become a trusted partner of the FOPH, demonstrating the importance of genomic data curation and management to efficiently support national surveillance programs. It is essential that such infrastructures are consolidated to support current and future surveillance programs.”
Katrin Schneider
Head Bioinformatics and Data Science Unit at the FOPH
Launched during the COVID-19 pandemic and mandated by the Federal Office of Public Health (FOPH) in 2021, SPSP was initially focused on SARS-CoV-2. Its mission has since expanded to include pathogens of national concern, such as influenza, respiratory syncytial virus (RSV), Listeria monocytogenes, and Salmonella, through collaboration with both the FOPH and the Federal Food Safety and Veterinary Office (FSVO).
Thanks to SPSP, Switzerland was ranked in the top5 contributing countries for SARS-CoV-2 sequences during the pandemic.

Benefits for Switzerland
SPSP serves two core objectives:
1. Surveillance
SPSP provides near real-time genomic surveillance of pathogens across human and veterinary medicine, including environmental and food sources. It enables public health authorities to:
-
- accelerate research on pathogens;
- detect pandemics early and respond decisively;
- monitor transmission dynamics more effectively;
- identify new variants and resistant strains;
- make surveillance funding more impactful.
- generate actionable insights through tools such as Nextstrain.
Pipelines include variant calling, outbreak detection, and phylogenetic visualization, supporting timely and informed public health responses.
2. Research
SPSP is also a trusted national data repository for infectious disease research. Its high-quality, well-annotated datasets:
-
- adhere to common standards;
- are interoperable with Swiss BioMedIT;
- enable advanced search, analysis, and integration;
- can be assigned DOIs to ensure reproducibility and traceability.
Benefits for global pathogen surveillance
Pathogen genomic surveillance is a cornerstone of modern public health. Through SPSP, Switzerland contributes to international efforts such as the WHO’s International Pathogen Surveillance Network (IPSN) and the Pathogens Portal —helping strengthen global surveillance systems and preparedness.
“The IPSN Community of Practice on Genomics Data is an important mechanism to improve global pathogen data practices and sharing.”
Aitana Neves
Deputy Chair of SPSP
Member of IPSN’s Community of Practice on Genomics Data
Associate Director, Clinical Bioinformatics group, SIB
Benefits for data providers and users
SPSP centralizes and harmonizes data, offering broad utility for clinical labs, public health agencies, and researchers:
-
- a single entry point for sequencing data from diverse sources;
- common data standards that ensure comparability across time and regions;
- curation and validation before data is reused or shared externally;
- secure de-identification methods and privacy-compliant access;
- custom reports tailored to public health needs;
- ability to link genomes across hosts and environments for deeper insight.